
In 1969 Cyrus Levinthal noted that it would take longer than the age of the known universe to enumerate all possible configurations of a typical protein by brute force calculation – Levinthal estimated 10^300 possible conformations for a typical protein. A major challenge, however, is that the number of ways a protein could theoretically fold before settling into its final 3D structure is astronomical. This hypothesis sparked a five decade quest to be able to computationally predict a protein’s 3D structure based solely on its 1D amino acid sequence as a complementary alternative to these expensive and time consuming experimental methods. In his acceptance speech for the 1972 Nobel Prize in Chemistry, Christian Anfinsen famously postulated that, in theory, a protein’s amino acid sequence should fully determine its structure. These techniques, as well as newer methods like cryo-electron microscopy, depend on extensive trial and error, which can take years of painstaking and laborious work per structure, and require the use of multi-million dollar specialised equipment.

This has been a focus of intensive scientific research for many years, using a variety of experimental techniques to examine and determine protein structures, such as nuclear magnetic resonance and X-ray crystallography. - Professor John Moult, Co-founder and Chair of CASP, University of Maryland To see DeepMind produce a solution for this, having worked personally on this problem for so long and after so many stops and starts, wondering if we’d ever get there, is a very special moment. We have been stuck on this one problem – how do proteins fold up – for nearly 50 years. Many of the world’s greatest challenges, like developing treatments for diseases or finding enzymes that break down industrial waste, are fundamentally tied to proteins and the role they play. This breakthrough demonstrates the impact AI can have on scientific discovery and its potential to dramatically accelerate progress in some of the most fundamental fields that explain and shape our world.Ī protein’s shape is closely linked with its function, and the ability to predict this structure unlocks a greater understanding of what it does and how it works.

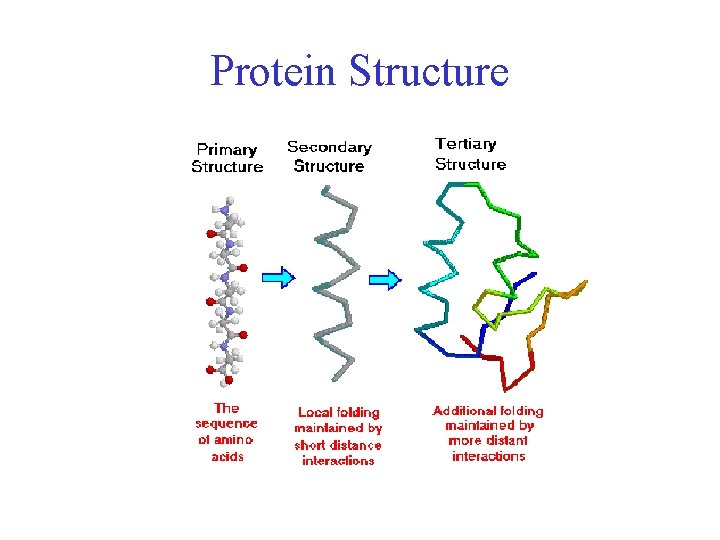
In a major scientific advance, the latest version of our AI system AlphaFold has been recognised as a solution to this grand challenge by the organisers of the biennial Critical Assessment of protein Structure Prediction ( CASP ). Figuring out what shapes proteins fold into is known as the “protein folding problem”, and has stood as a grand challenge in biology for the past 50 years. They are large complex molecules, made up of chains of amino acids, and what a protein does largely depends on its unique 3D structure. Proteins are essential to life, supporting practically all its functions.
In July 2022, we released AlphaFold protein structure predictions for nearly all catalogued proteins known to science.


 0 kommentar(er)
0 kommentar(er)
